MPAS unstructured grid data#
In this example, we demonstrate CE identification with unstructured-grid data from MPAS
and compare to the results using the regridded dataset.
import warnings
import cartopy
import cartopy.crs as ccrs
import cartopy.feature as cfeature
import matplotlib as mpl
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import xarray as xr
from matplotlib.tri import Triangulation
import tams
warnings.filterwarnings("ignore", category=cartopy.io.DownloadWarning)
%matplotlib inline
xr.set_options(display_expand_data=False)
Show code cell output
ERROR 1: PROJ: proj_create_from_database: Open of /home/docs/checkouts/readthedocs.org/user_builds/tams/conda/latest/share/proj failed
<xarray.core.options.set_options at 0x7f4d0191d0f0>
Load data#
ds = tams.load_example_mpas_ug()
ds
<xarray.Dataset>
Dimensions: (time: 127, cell: 133447)
Coordinates:
* time (time) datetime64[ns] 2006-09-08T12:00:00 ... 2006-09-13T18:00:00
lat (cell) float64 ...
lon (cell) float64 ...
Dimensions without coordinates: cell
Data variables:
tb (time, cell) float32 nan nan nan nan ... 264.8 264.6 266.4 263.8
precip (time, cell) float32 0.0323 0.03974 0.0 0.1706 ... nan nan nan nanfig, ax = plt.subplots(figsize=(6, 3))
sel = ds.isel(cell=slice(None, None, 20))
ax.scatter(sel.lon, sel.lat, marker=".", s=10, alpha=0.5, edgecolors="none")
ax.set(xlabel="lon", ylabel="lat")
ax.autoscale(tight=True)
%%time
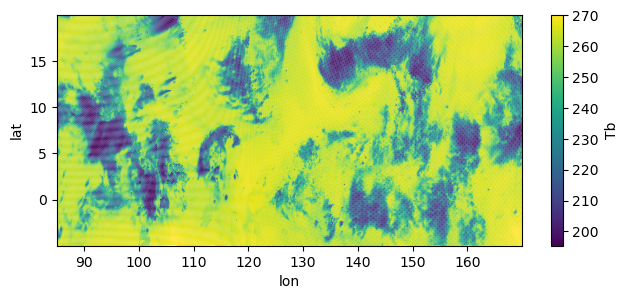
fig, ax = plt.subplots(figsize=(7.5, 3))
im = ax.scatter(ds.lon, ds.lat, c=ds.tb.isel(time=10), marker=".", s=3, edgecolors="none")
fig.colorbar(im, ax=ax, label="Tb")
ax.autoscale(tight=True)
ax.set(xlabel="lon", ylabel="lat")
CPU times: user 91.6 ms, sys: 0 ns, total: 91.6 ms
Wall time: 90.6 ms
[Text(0.5, 0, 'lon'), Text(0, 0.5, 'lat')]
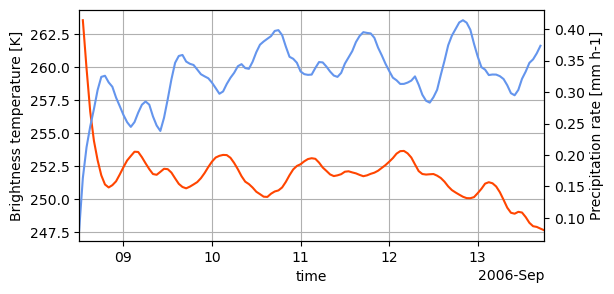
ts = ds.mean("cell", keep_attrs=True)
fig, ax = plt.subplots(figsize=(6, 3))
ax2 = ax.twinx()
ts.tb.plot(ax=ax, c="orangered")
ts.precip.plot(ax=ax2, c="cornflowerblue")
ax.autoscale(axis="x", tight=True)
ax.grid(True)
Identify CEs#
%%time
itime = 10
stime = pd.Timestamp(ds.time.values[itime]).strftime(r"%Y-%m-%d_%H")
print(stime)
x = ds.lon
y = ds.lat
tri = Triangulation(x, y)
2006-09-08_22
CPU times: user 1.01 s, sys: 36 ms, total: 1.04 s
Wall time: 1.04 s
%%time
# Passing the triangulation in is not required but makes it faster
shapes = tams.core._contours_to_gdf(tams.contours(ds.tb.isel(time=itime), value=235, triangulation=tri));
CPU times: user 295 ms, sys: 3.62 ms, total: 299 ms
Wall time: 867 ms
%%time
# `tams.identify` does the above but also for the core threshold and does size filtering by default
cs_ug, cs_ug_core = list(zip(*tams.identify(ds.tb.isel(time=10))))[0]
CPU times: user 1.93 s, sys: 83.6 ms, total: 2.01 s
Wall time: 2.01 s
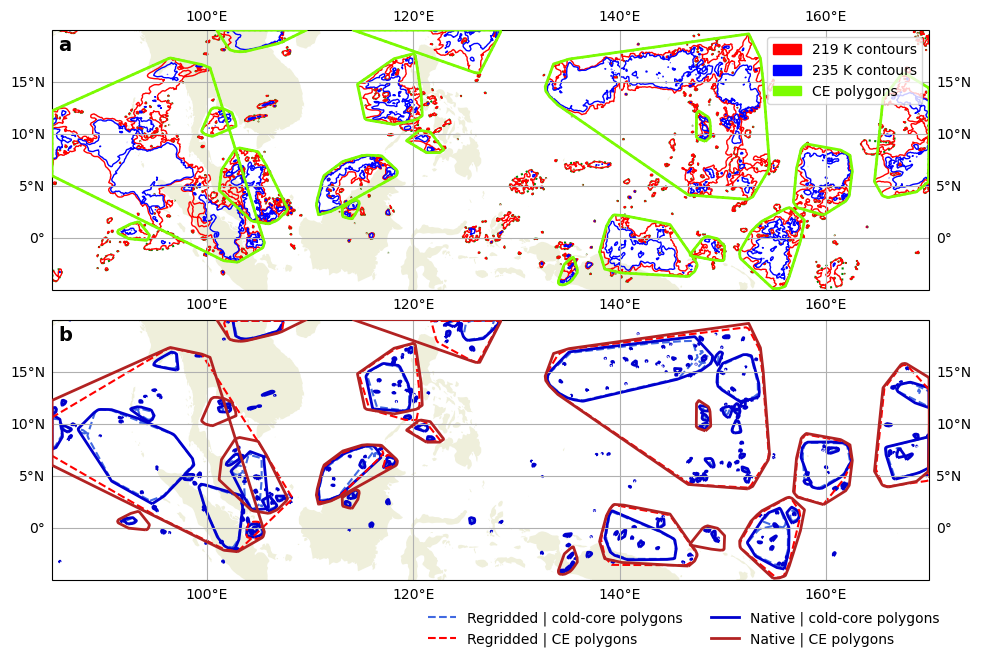
%%time
tran = ccrs.PlateCarree()
proj = ccrs.PlateCarree() # near equator
fig, (ax1, ax2) = plt.subplots(2, 1, figsize=(10, 6), subplot_kw=dict(projection=proj), constrained_layout=True)
ax = ax1
ax.add_feature(cfeature.LAND)
tcs = ax.tricontour(tri, ds.tb.isel(time=itime), levels=[235], colors="red", linewidths=1, transform=tran)
tcs_core = ax.tricontour(tri, ds.tb.isel(time=itime), levels=[219], colors="blue", linewidths=1, transform=tran)
shapes.plot(fc="none", ec="green", lw=1.5, ls=":", ax=ax, transform=tran) # not size-filtered
cs_ug.plot(fc="none", ec="lawngreen", lw=2, ax=ax, zorder=3, transform=tran)
ax.gridlines(draw_labels=True)
legend_handles = [
mpl.patches.Patch(color="red", label="219 K contours"),
mpl.patches.Patch(color="blue", label="235 K contours"),
mpl.patches.Patch(color="lawngreen", label="CE polygons"),
]
ax.legend(handles=legend_handles, loc="upper right")
ax = ax2
ds_rg = tams.load_example_mpas().sel(lat=slice(None, 20)) # same lat upper bound as in the ug data
ax.add_feature(cfeature.LAND)
cs_rg, cs_rg_core = list(zip(*tams.identify(ds_rg.tb.isel(time=itime))))[0]
a = cs_rg_core.plot(fc="none", ec="royalblue", lw=1.5, ls="--", transform=tran, ax=ax)
cs_ug_core.plot(fc="none", ec="mediumblue", lw=2, transform=tran, ax=ax)
cs_rg.plot(fc="none", ec="red", lw=1.5, ls="--", transform=tran, ax=ax)
cs_ug.plot(fc="none", ec="firebrick", lw=2, zorder=3, transform=tran, ax=ax)
legend_handles = [
mpl.lines.Line2D([], [], color="royalblue", ls="--", lw=1.5, label="Regridded | cold-core polygons"),
mpl.lines.Line2D([], [], color="red", ls="--", lw=1.5, label="Regridded | CE polygons"),
mpl.lines.Line2D([], [], color="mediumblue", ls="-", lw=2, label="Native | cold-core polygons"),
mpl.lines.Line2D([], [], color="firebrick", ls="-", lw=2, label="Native | CE polygons"),
]
gl = ax.gridlines(draw_labels=True)
gl.top_labels = False
# ax.autoscale(tight=True)
ax.set_xlim(ax1.get_xlim()); ax.set_ylim(ax1.get_ylim())
fig.legend(handles=legend_handles, ncol=2, loc="lower right", bbox_to_anchor=[0, -0.09, 0.961, 1], frameon=False)
for a, ax in zip("abc", [ax1, ax2]):
ax.text(0.007, 0.98, a, weight="bold", size=14, va="top", ha="left", transform=ax.transAxes)
fig.savefig(f"mpas-ug-contours-and-vs-rg-ces_{stime}.pdf", bbox_inches="tight", pad_inches=0.05, transparent=False)
CPU times: user 2.65 s, sys: 35.9 ms, total: 2.69 s
Wall time: 3.14 s
Triangulations#
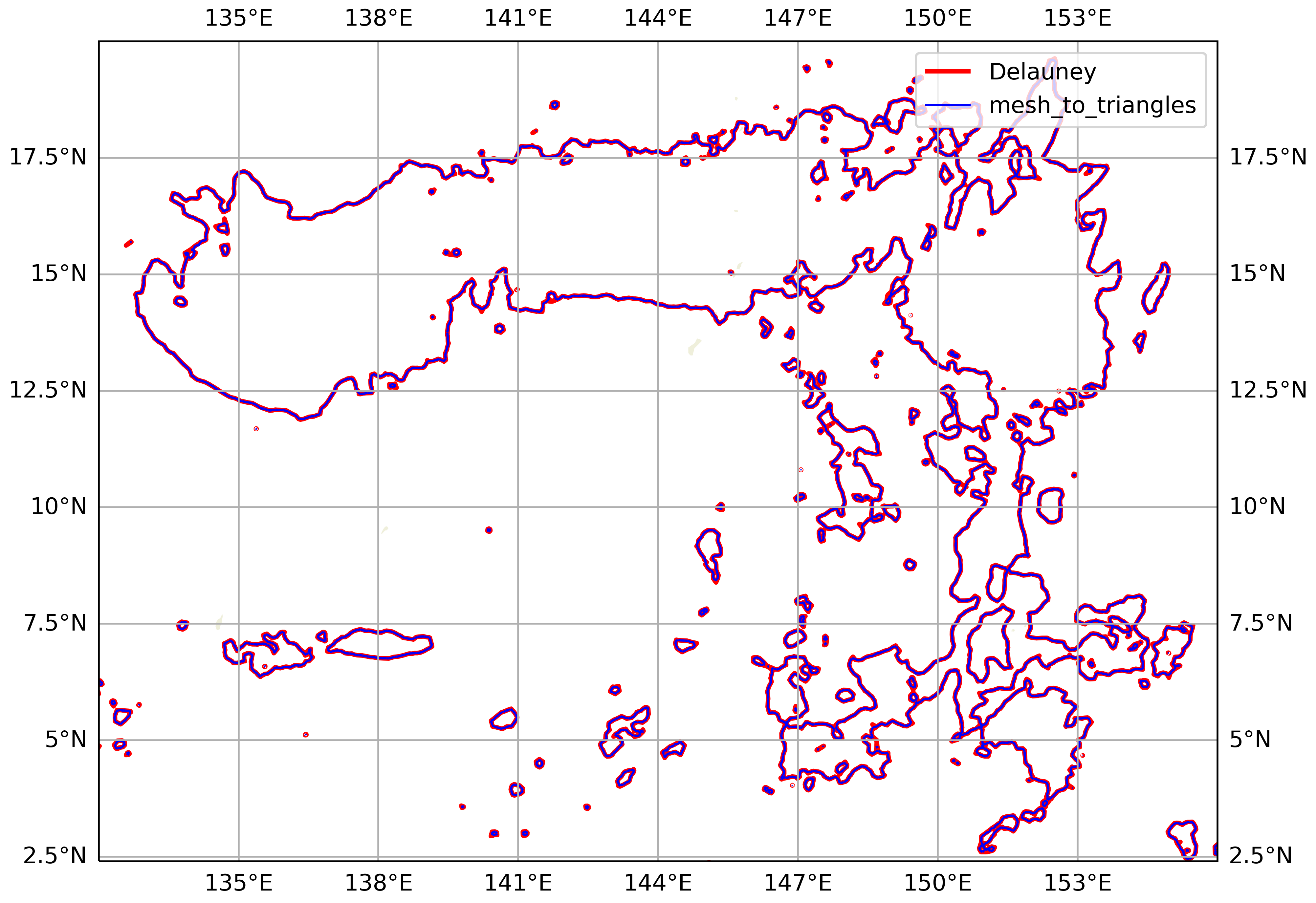
The above contouring is based on the Delauney triangulation. This connects the cell centers with straight lines to form triangles. But this triangulation doesn’t correspond to the MPAS mesh, per se.
MPAS-Tools, though, provides a method for dividing the mesh grid cells into triangles and interpolating data defined at cell centers (like OLR) to the triangle nodes. The triangle nodes include the cell centers (one per triangle) and the grid cell vertices (two per triangle). A hexagonal grid cell, e.g., is divided into 6 triangles, which all share the cell center as a node.
We compared the contourings from these two methods. For our intents and purposes (identifying CEs with area ≥ 4000 km²), the differences appear to be negligible. The mesh horizontal resolution is 15 km, which seems to be sufficiently high that the Delauney triangulation is a good approximation.
# Code